Others
Find the netlist corresponding to param
1. Find a set of params that succeeded during the validation process
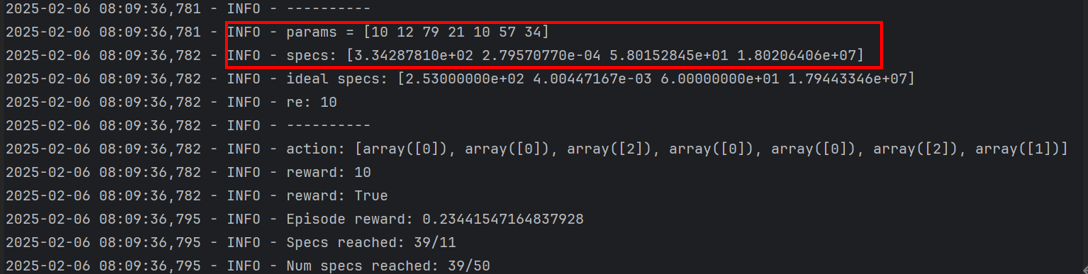 Params = [10 12 79 21 10 57 34]
Params = [10 12 79 21 10 57 34]
2. The params output here is index, and the actual value is calculated according to the specification range in two_stage_opamp.yaml
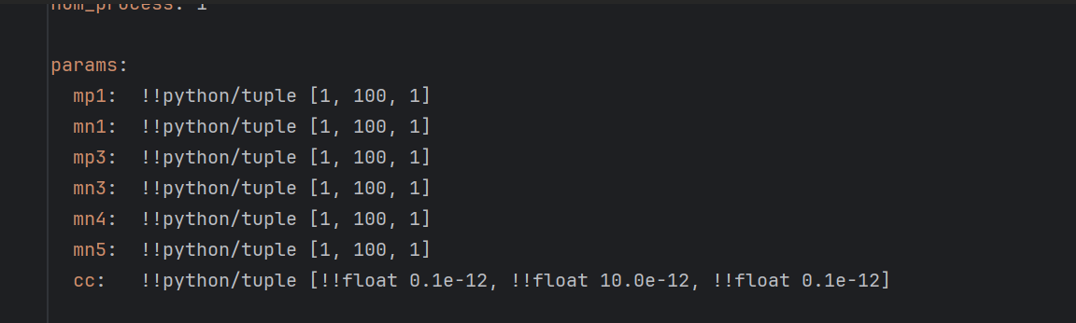 The first 6 are: $1 + 1 \times x$, the last one is: $(0.1 + 0.1 \times x)\times 10^{-12}$
The first 6 are: $1 + 1 \times x$, the last one is: $(0.1 + 0.1 \times x)\times 10^{-12}$
So the final params = [11 13 80 22 11 58 3.5e-12]
3. According to ngspice_wrapper.py, the changed netlist exists in the BASE_TMP_DIR path:
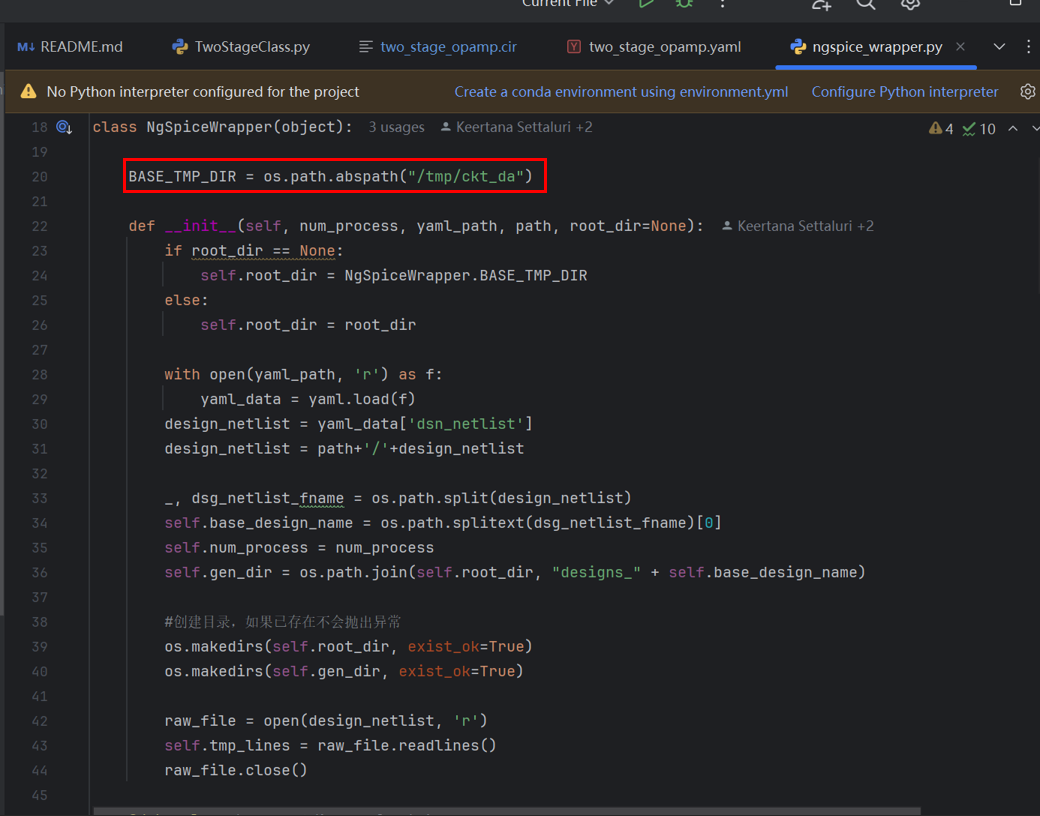
4. Enter /tmp/ckt_da/designs_two_stage_opamp/, enter cd two_stage_opamp_ + the params just calculated, and you can enter the corresponding path:
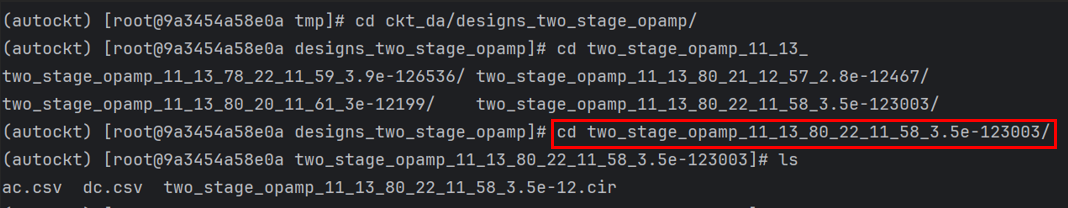
The Cir file is opened to get the netlist parameters. Its file name and path name are generated using the actual value of params.
Code error location (ac/dc file doesn’t exist)
In the file AutoCkt/eval_engines/ngspice/TwoStageClass.py around line 47, the following code checks for the existence of .ac and .dc simulation results:
if not os.path.isfile(ac_fname) or not os.path.isfile(dc_fname):
log.warning("ac/dc file doesn't exist: {}".format(output_path))
This warning is often triggered when the simulation process fails to generate the required files. Here are the common causes and how to resolve them:
Reason 1: $SPECTRE_MODEL_PATH Not Set
The spectre log will show error as: ./design_wrapper.lib.scs not exist or cannot find ./design_wrapper.lib.scs.
If your netlist includes a model like:
include "$SPECTRE_MODEL_PATH/design_wrapper.lib.scs" section=mainlib
You need to ensure the environment variable SPECTRE_MODEL_PATH is correctly set before running the code.
If not, Spectre will fail silently, and the .ac/.dc files will not be generated.
Fix: Add the following line in your C shell environment (csh):
setenv SPECTRE_MODEL_PATH /net/vlsiserver/usr11/library/CSM/gf55BCDlite_v1090_stack7A/55BCDlite/V1.0_9.0/Models/Spectre/55bcdlite/models/
Reason 2: Wrong Shell – Must Use csh
This codebase requires activating the Spectre simulation environment via conda in a C shell (csh).
Running the script in bash will not activate the correct environment and lead to simulation failure.
Fix: Always launch your terminal with csh before activating conda and running the scripts.
Reason 3: Simulation Command Fails Silently
The simulation is performed in eval_engines/ngspice/ngspice_wrapper.py (lines 117–123) using this command block:
command = """
spectre "{0}" -o "{1}" -log >& /dev/null
ocean -nograph -restore "{2}" >& /dev/null
find "{1}" -name ".*.dep" -exec rm -rf +
find "{1}" -name "*.raw" -exec rm -rf +
""".format(fpath, output_dir, os.path.join(output_dir, "export.ocn"))
Note that both spectre and ocean logs are hidden by redirecting output to /dev/null.
This means if any error occurs, you won’t see it by default.
Fix:
-
Run
spectreandoceancommands manually using the netlist and.ocnscript to check:- If the netlist has syntax or model errors
- If the ocean script is written correctly
- If all required files exist in the specified output directory
✅ Summary
If you see ac/dc file doesn't exist:
- Check that
SPECTRE_MODEL_PATHis set in your environment. - Confirm you’re using
cshshell with the correct conda environment. - Debug Spectre and Ocean commands manually to check for hidden errors.
This step is essential to ensure correct simulation output before proceeding with reinforcement learning training.
❗ Error: “- WARNING - results file doesn’t exist”
This warning typically appears when the result.txt file was not successfully generated after running simulation.
🔍 Root Cause
The most common reason is that Cadence log files (e.g., CDS.log) are already occupied by other processes. As a result, the ocean script fails to write logs properly and skips generating the result.txt file.
To confirm this, check the contents of ocean.log — it will usually show signs of resource conflict or failure to write logs.
🛠️ Solution
-
Use the following command to forcefully kill leftover Python processes that might be occupying the logs:
pkill -9 -f python -
After that, restart the conda environment and run the code again.